Effect of intracellular environment on enzyme function
Advisor: Štěpán Timr (JH-INST CAS)
Funding: Fully funded
Website: http://www.stepantimr.com
Contact: stepan.timr@jh-inst.cas.cz
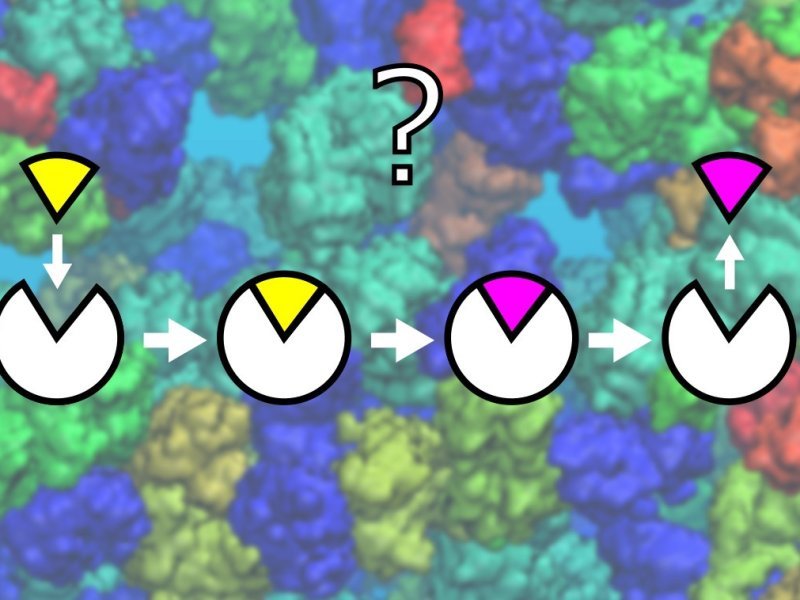
Metabolic reactions take place in a highly crowded cellular interior, which is filled with proteins, nucleic acids, metabolites, and other biological molecules. How interactions with such a dense and heterogeneous environment influence—and potentially control—enzyme activity, has not been fully elucidated yet. Understanding these effects would benefit from detailed atomistic insights provided by molecular simulations. The goal of this PhD work will be to link the composition and structure of the local environment around an enzyme to key factors determining enzyme activity, such as the conformational state of the enzyme molecule and binding preferences of its substrates. To reach this goal, the PhD student will establish a computational protocol combining enhanced-sampling molecular simulations with machine-learning approaches. He/she will apply the protocol to systems of growing complexity, ranging from experimentally thoroughly-characterized model systems to complex models of biomolecular condensates involving intrinsically disordered proteins and/or RNA. The results of the work will form an important building block of a multi-scale description of dynamic enzyme assemblies and their role in the regulation of cellular metabolism.