Computational modeling of dynamic enzyme assemblies
Advisor: Štěpán Timr (JH-INST CAS)
Funding: Fully funded
Website: http://www.stepantimr.com
Contact: stepan.timr@jh-inst.cas.cz
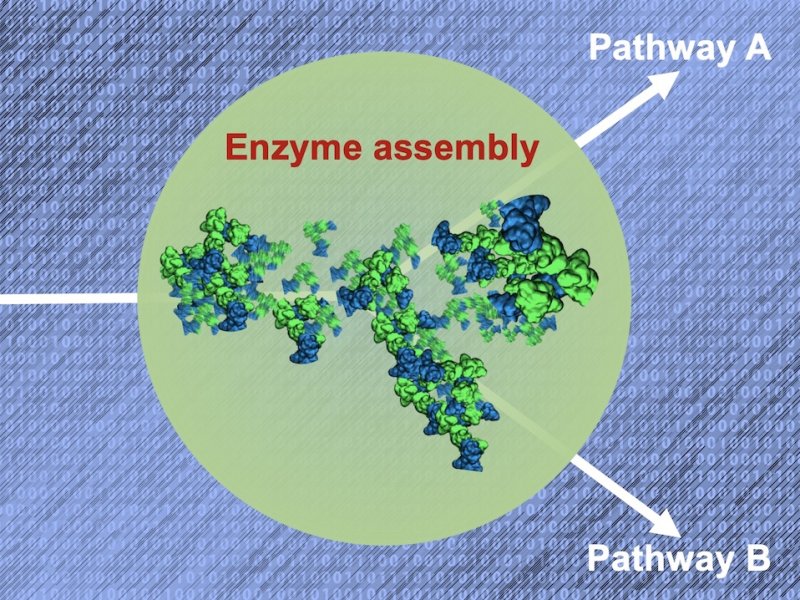
In living cells, various enzymes have been found to assemble into transient structures that can appear and disassemble as a function of external conditions. Among other examples, such dynamic assemblies have been identified in glycolysis or in the purine synthesis pathway. Recent experimental evidence points to a key role of dynamic enzyme assemblies in the regulation and adaptation of cellular metabolism, including their possible role as a switch between two or more competing pathways. However, the mechanisms underlying the formation and function of these assemblies are yet to be elucidated. By using a combination of atomistic, coarse-grained and ultra-coarse-grained molecular modeling and working in tight connection with experimental data, the PhD student will characterize molecular interactions promoting assembly formation and quantify the diffusivities of enzymes and reactants inside dynamic enzyme assemblies. In particular, he/she will evaluate the potential for substrate channeling, that is, passing the intermediate products efficiently between consecutive enzymes of a pathway. The computational methodology developed in this work will serve as a basis for the prediction of metabolic fluxes given a composition and architecture of an enzyme assembly.